
Atomistic Simulations and In Silico Mutational Profiling of Protein Stability and Binding in the SARS-CoV-2 Spike Protein Complexes with Nanobodies: Molecular Determinants of Mutational Escape Mechanisms
5
(265) ·
$ 4.99 ·
In stock
Description

Atomistic Simulations and Deep Mutational Scanning of Protein Stability and Binding Interactions in the SARS-CoV-2 Spike Protein Complexes with Nanobodies: Molecular Determinants of Mutational Escape Mechanisms

IJMS, Free Full-Text

Atomistic Simulations and In Silico Mutational Profiling of Protein Stability and Binding in the SARS-CoV-2 Spike Protein Complexes with Nanobodies: Molecular Determinants of Mutational Escape Mechanisms

Plausible blockers of Spike RBD in SARS-CoV2—molecular design and underlying interaction dynamics from high-level structural descriptors

In silico investigation on the mutational analysis of BRCA1-BARD1 RING domains and its effect on nucleosome recognition and ubiquitination - ScienceDirect
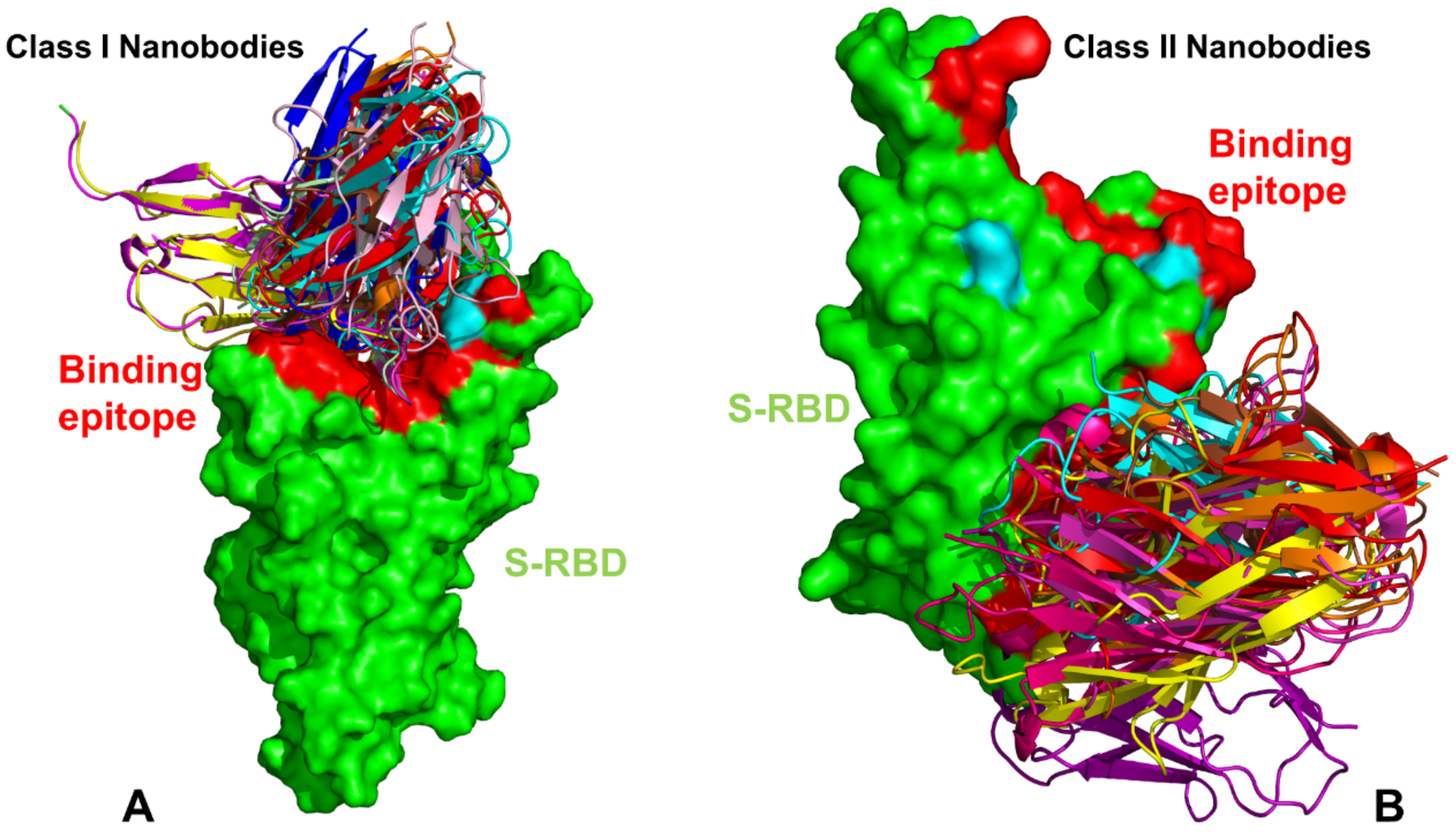
IJMS, Free Full-Text
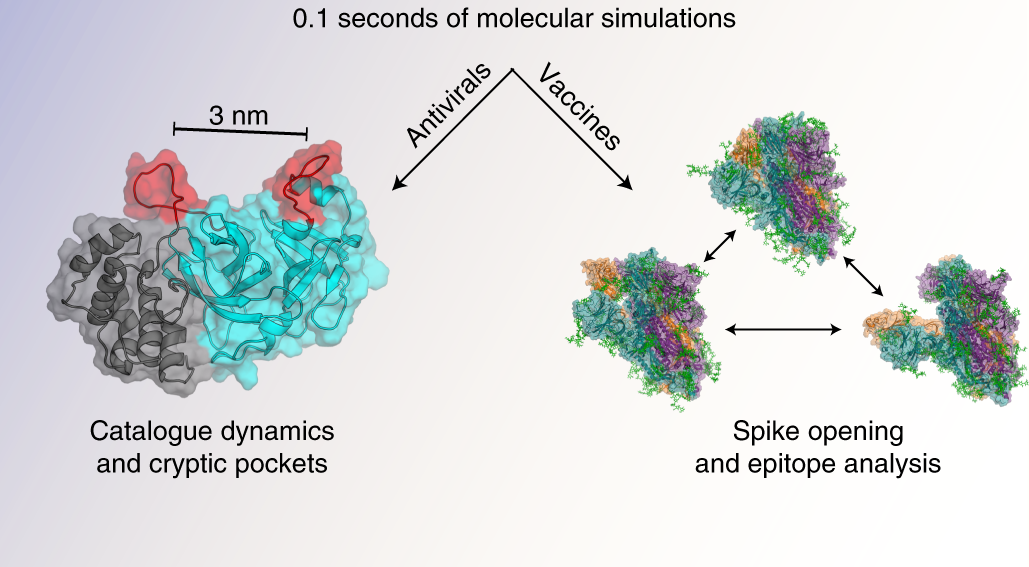
SARS-CoV-2 simulations go exascale to predict dramatic spike opening and cryptic pockets across the proteome
In silico analysis of SARS-CoV-2 spike glycoprotein and insights into antibody binding

Impact of new variants on SARS-CoV-2 infectivity and neutralization: A molecular assessment of the alterations in the spike-host protein interactions - ScienceDirect
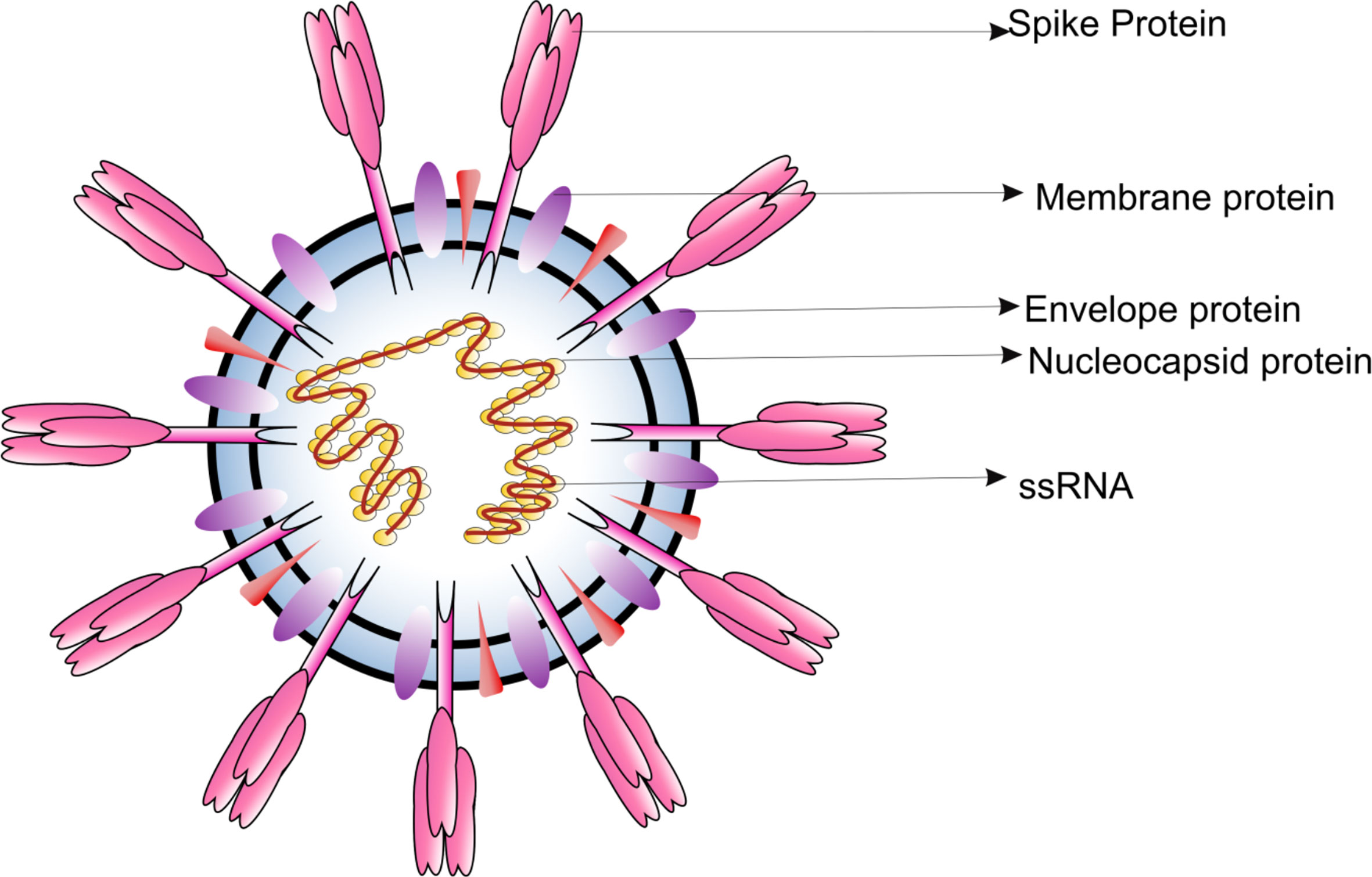
Frontiers Potential Therapeutic Targets and Vaccine Development for SARS- CoV-2/COVID-19 Pandemic Management: A Review on the Recent Update

Highly synergistic combinations of nanobodies that target SARS-CoV-2 and are resistant to escape
Related products
You may also like
copyright © 2019-2024 osihenoutlet.com all rights reserved.








